一、进行GO富集分析
网站:
agriGO: GO Analysis Toolkit and Database for Agricultural Community
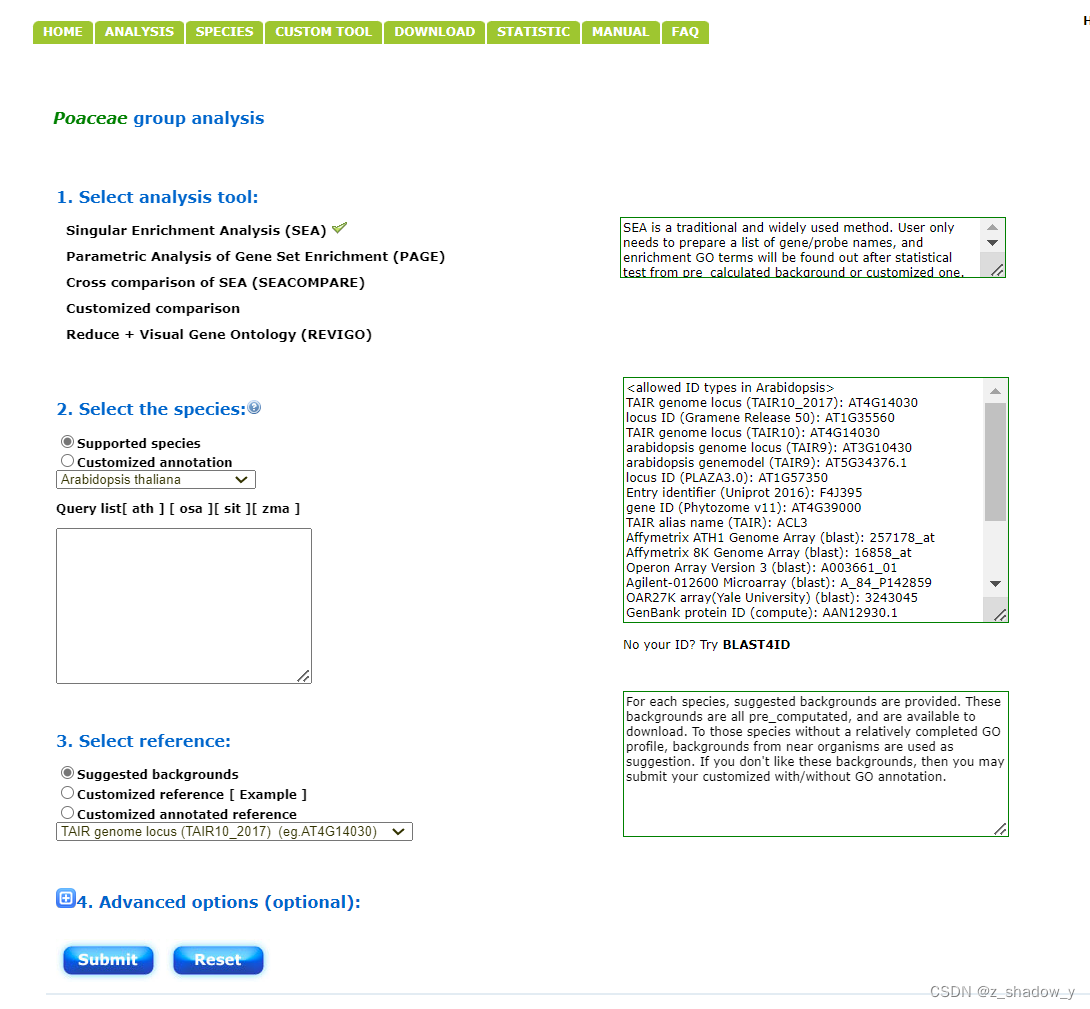
提供geneid即可进行分析
结果如下:
 注意: download是下载所有结果;仅选取FDR<0.05的结果下载即可
注意: download是下载所有结果;仅选取FDR<0.05的结果下载即可
二、可视化——GOplot
所需文件:1、富集结果 2、差异基因的表达差异倍数文件
(一)处理文件格式
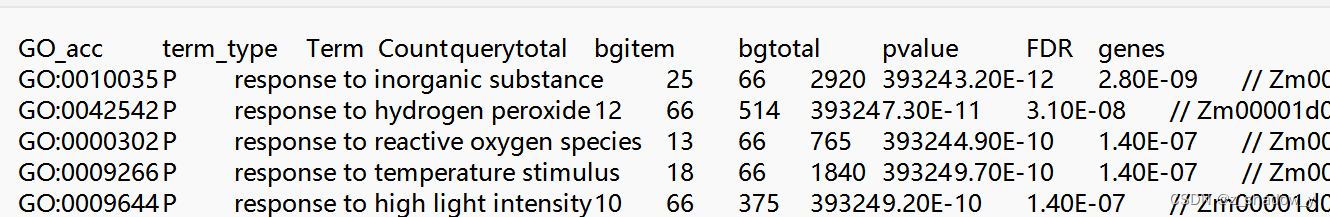
1、富集结果——D:\张颖\other\DM\GO\GO_5to8_result.csv

goenrichment <- read.csv(file = "clipboard",header = T,sep = "\t") #读入GO富集分析的结果
goenrichment <- goenrichment[ , c(3,1,2,7,4)] #按照要求的顺序选择列
colnames(goenrichment) <- c("category", "ID", "term", "adj_pval", "genes")
goenrichment$genes <- gsub(";", ",", goenrichment$genes) # 用逗号替换分号
2、差异基因文件——D:\张颖\other\DM\GO\2genefile.csv
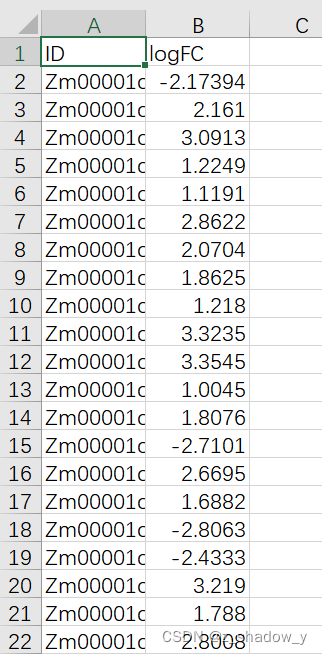
D:\张颖\other\DM\GO\make_2genefile.py
#制作GO_5to8_result文件中涉及基因的差异表达文件
#ID logFC
import os
# dir=os.getcwd()
# print(dir) 就在GO目录下
gene_list=[]
data=open("GO_5to8_result.csv","r")
for line in data.readlines():
genes=line.replace("\n","").split(",",4)[4]
gene_list.extend(genes.split(","))
data.close()
dic={}
DGE=open("8L_27_24vsH_27_24.DEG.CSV","r")
for l in DGE.readlines():
id=l.split(",")[0]
logFC=l.split(",")[3]
if id in gene_list:
dic[id]=logFC
#print(dic)
DGE.close()
output=open("2genefile.csv","a+")
for k in dic.keys():
output.write(k+","+dic[k]+"\n")
output.close()
(二)绘图
D:\张颖\other\DM\GO\GOplot.R
#GO富集分析弦图绘制
rm(list = ls())
#install.packages("GOplot")
library(GOplot)
library(viridis)
data1="GO_5to8_result.csv"
data2="2genefile.csv"
d1 <- read.csv(data1, sep=',',header=T)
d2 <- read.csv(data2, sep=',',header=T)
circ=circle_dat(d1,d2)
d2$ID=toupper(d2$ID)
chord=chord_dat(circ,d2,d1$Term)pdf("chord.pdf",height = 13,width = 13)
GOChord(chord,space = 0.02,#GO term处间隔大小设置
gene.order = "logFC",
limit = c(5, 7),
gene.space = 0.26,gene.size = 2.8,
process.label = 7,
ribbon.col=viridis_pal(option = "D")(23)
#ribbon.col=colorRampPalette(brewer.pal(12,"Set3"))(23)
)
dev.off()
注意事项:


参考:
https://www.jianshu.com/p/1a686b376821
三、修改图时的重难点
1、颜色设置
GOChord(chord, title="GOChord plot",#标题设置 space = 0.02, #GO term处间隔大小设置 limit = c(3, 5),#第一个数值为至少分配给一个基因的Goterm数,第二个数值为至少分配给一个GOterm的基因数 gene.order = 'logFC', gene.space = 0.25, gene.size = 5,#基因排序,间隔,名字大小设置 lfc.col=c('firebrick3', 'white','royalblue3'),##上调下调颜色设置 #ribbon.col=colorRampPalette(c("blue", "red"))(length(EC$process)),#GO term 颜色设置 ribbon.col=brewer.pal(length(EC$process), "Set3"),#GO term 颜色设置
提前跑一下看一下GOterm的数目,再设置color的数目,否则会报错
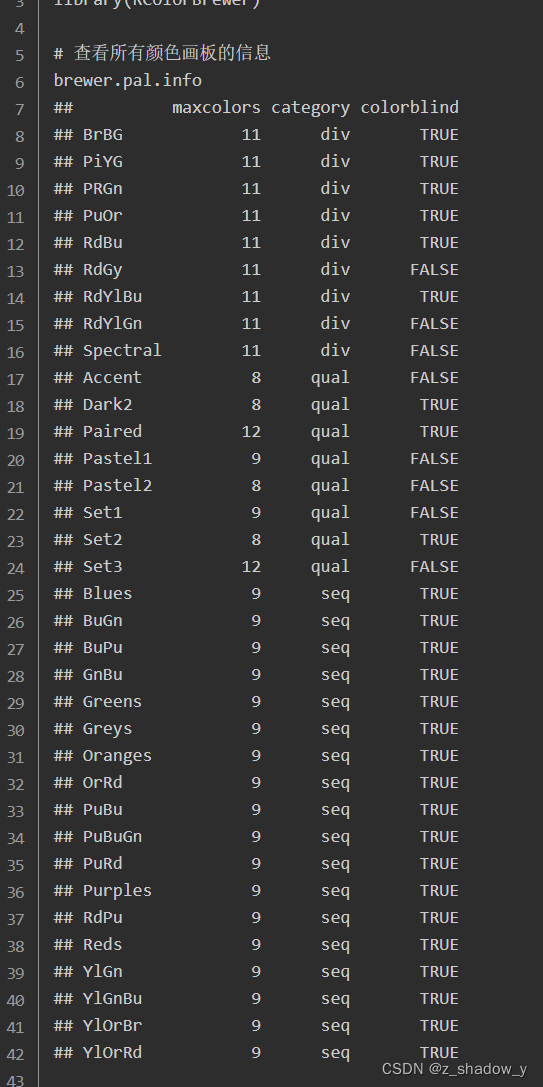
2、颜色上限
R的常用配色包:
https://www.jianshu.com/p/b1897f06328d
四、课余学习
GO富集可视化-GOplot | KeepNotes blog
气泡图
五、绘制气泡图
library(ggplot2)
library(dplyr)
GO_dataset=read.table(file="GO_5to8_result.txt",
header = TRUE,
sep = "\t")
GO_dataset=arrange(GO_dataset,GO_dataset[,8])
GO_dataset$Term=factor(GO_dataset$Term,
levels = rev(GO_dataset$Term))
###图片背景设定
mytheme <- theme(axis.title=element_text(face="bold", size=14,colour = 'black'), #坐标轴标题
axis.text=element_text(face="bold", size=14,colour = 'black'), #坐标轴标签
axis.line = element_line(size=0.5, colour = 'black'), #轴线
panel.background = element_rect(color='black'), #绘图区边框
legend.key = element_blank() #关闭图例边框
)
p <- ggplot(GO_dataset,aes(x=Count,y=Term,colour=-1*log10(pvalue),size=Count))+
geom_point()+
scale_size(range=c(6, 10))+
scale_colour_gradient(low = "blue",high = "red")+
theme_bw()+
ylab("GO Pathway Terms")+
xlab("Gene Count")+
labs(color=expression(-log[10](pvalue)))+
theme(legend.title=element_text(size=8),
legend.text = element_text(size=6))+
theme(axis.title.y = element_text(margin = margin(r = 50)),
axis.title.x = element_text(margin = margin(t = 20)))+
theme(axis.text.x = element_text(face ="bold",color="black",angle=0,vjust=1,size = 6),
axis.text.y = element_text(face ="bold",color="black",angle=0,vjust=1,size = 6))
plot <- p+mytheme
plot
ggsave(plot,filename = "GO_5-8.pdf",width = 10,height = 6,dpi=300)
ggsave(plot,filename = "GO_5-8.png",width = 10,height = 6,dpi=300)
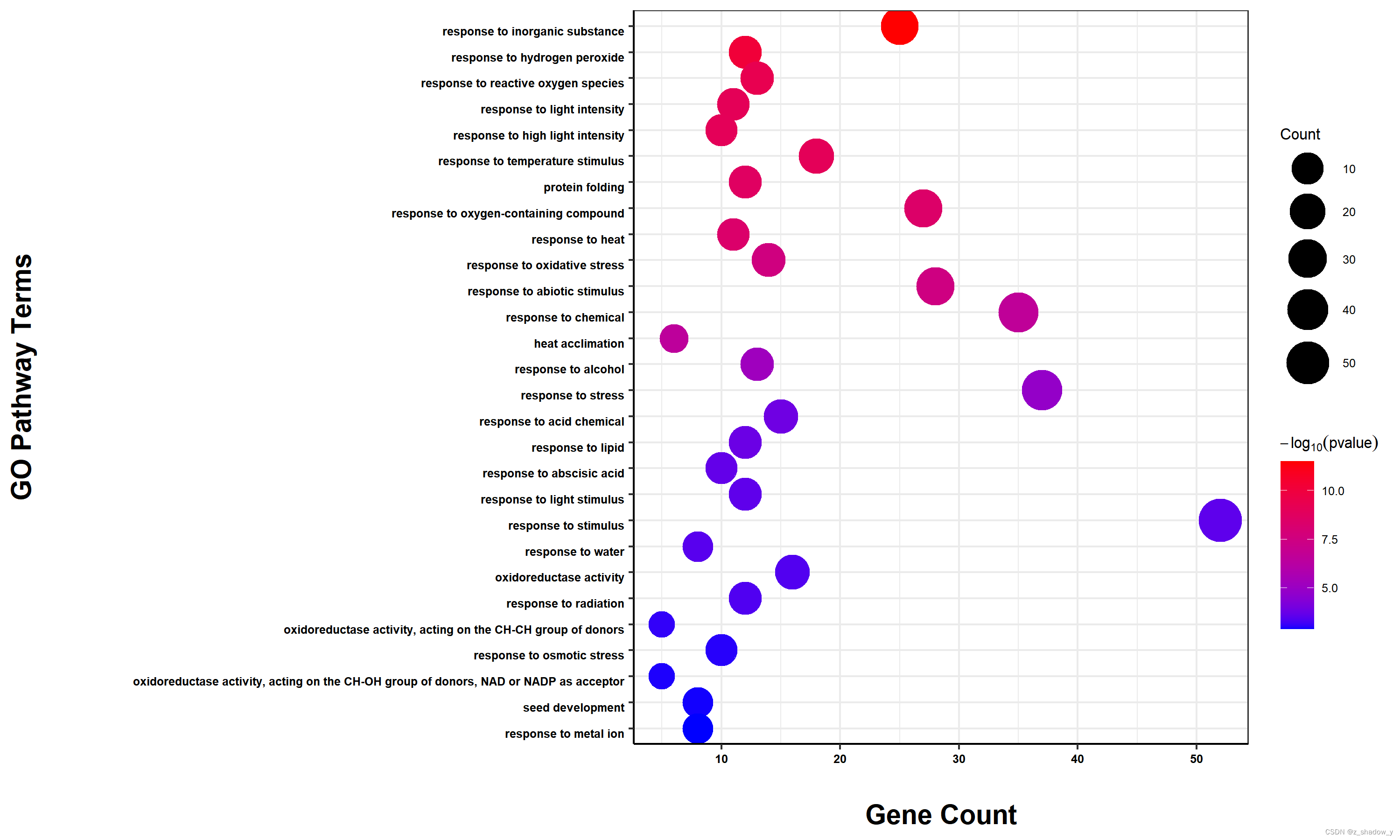
原始文件:
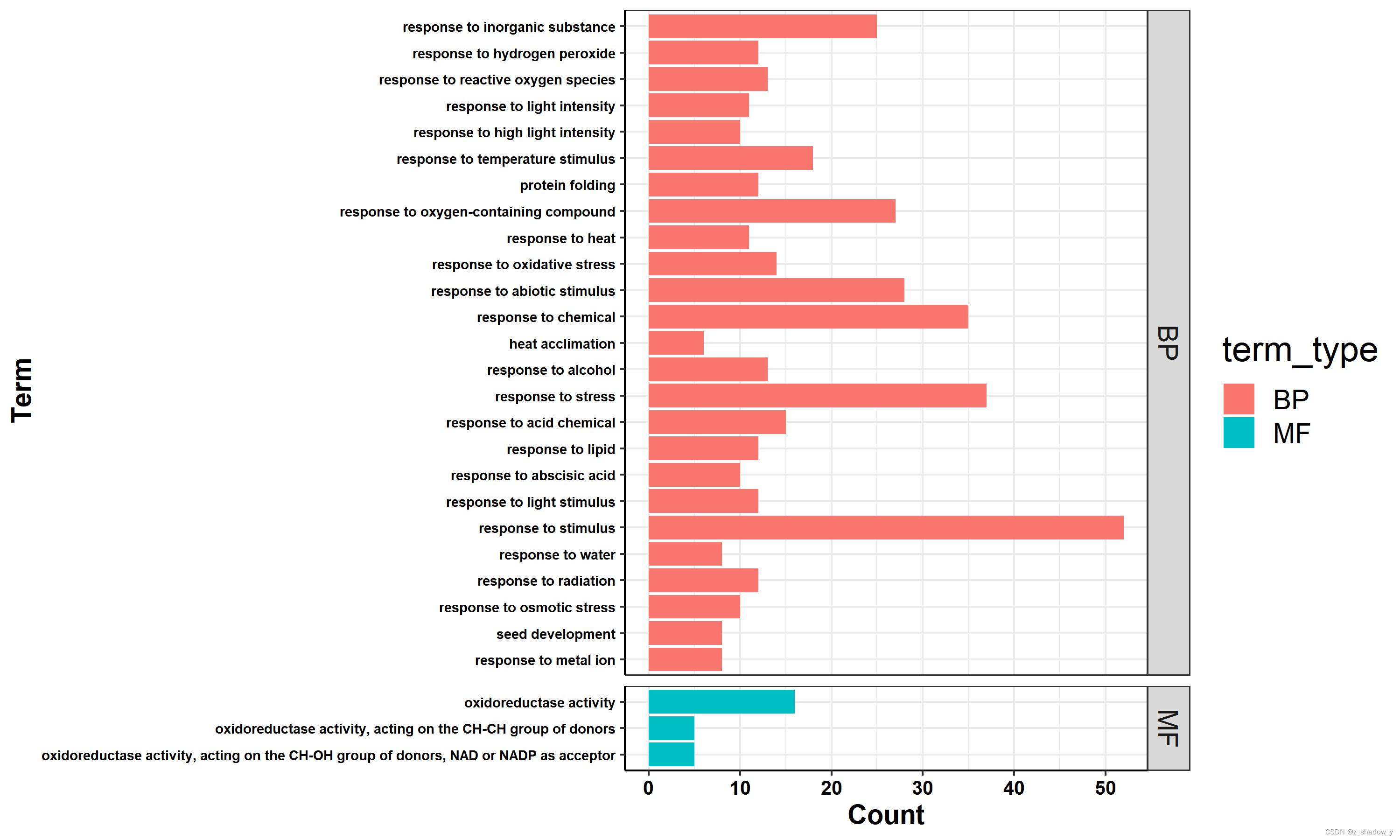
条形图
GO_dataset$term_type[which(GO_dataset$term_type=="F")]="MF"
p2 <- ggplot(GO_dataset,aes(y=Count,x=Term,fill=term_type)) +
geom_bar(stat="identity",position = "dodge") +
facet_grid(term_type~.,scales = "free",space = "free") +
coord_flip() +
theme_bw() +
theme(plot.title = element_text(hjust = 0.5),
strip.text.y = element_text(size = 14),
legend.position="right",
legend.title = element_text(size=18),
legend.text = element_text(size=14),
axis.text.x = element_text(size=10),
axis.text.y = element_text(size=7),
axis.title.x = element_text(size=14),
axis.title.y = element_text(size=14))
plot2 <- p2+mytheme
plot2